By Catherine Zandonella, Office of the Dean for Research
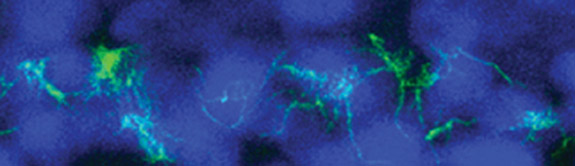
A new study of a protein found in cilia – the hair-like projections on the cell surface – may help explain how genetic defects in cilia play a role in developmental abnormalities, kidney disease and a number of other disorders.
The researchers at Princeton University and Northwestern University found that the protein, which goes by the name C21orf59 or “Kurly,” is needed for cilia to undulate to keep fluid moving over the surface of cells. They also found that the protein is needed during development to properly orient the cilia so that they are facing the right direction to move the fluid.
“It’s extremely exciting that we’ve found a single protein that is responsible for these two functions – orientation and motility – in cilia,” said Rebecca Burdine, an associate professor of molecular biology at Princeton University. “Despite their importance in human disease, very little is known about how cilia motility and orientation are coordinated, so this protein will provide an important gateway into looking at this process.” The finding is published online and in the March 1 issue of the journal Cell Reports.
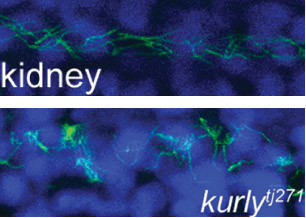
The studies were conducted in zebrafish at Princeton and in African clawed frogs (Xenopus laevis) at Northwestern. In the zebrafish kidney, the researchers found that the Kurly protein enabled cilia to orient themselves in a uniform direction, and most importantly, in the proper direction to facilitate the flow of fluid along the narrow channels in the kidney. In frogs, the cilia on skin cells help move fluid along the surface of the animal during its larval stage. In both cases, knocking out the gene for Kurly caused the cilia to orient incorrectly thereby losing their ability to move in the waving fashion that helps push fluid along.
The discovery of Kurly’s role in cilia movement and orientation stemmed from work in the Burdine lab on fetal organ development, specifically an investigation of mutations that alter the left-right asymmetric orientation of the heart. Such mutations can result in an organ that is working properly but is an exact mirror image of a normal heart. During a search for genes involved in this left-right patterning, the Burdine team discovered that mutations in a gene they called kur, which codes for the Kurly protein, were linked to errors in left-right orientation in zebrafish heart.
As the team investigated kur, they noted that the mutation also affected the function of cilia. It has been known for some time that cilia are important for a number of jobs, from sensing the environment to facilitating fluid flow, to ensuring that the lungs excrete inhaled contaminants. Cilia genetic defects are linked to a number of human diseases, including polycystic kidney disease, respiratory distress, hearing loss, infertility, and left-right patterning disorders such as the one Burdine studies.
Researchers in Burdine’s laboratory found that Kurly’s role in cilia movement stems from its ability to ensure proteins called dynein arms are correctly located in the cilia. When the researchers knocked out the kur gene, the dynein proteins failed to form in the proper location.
The finding that a single protein is involved in both movement and orientation is surprising, said co-first author Daniel Grimes, a postdoctoral research associate in the Burdine lab. “These are two aspects that are both required to generate fluid flow, and we’d like to know how they are linked molecularly. This work adds a new gene that aids this discovery.”
The gene for Kurly has also been detected in relation to human cilia disorders, so the work may have an impact on understanding the mechanisms of human disease, Grimes added. The researchers also found that the mutation they discovered rendered the Kurly protein sensitive to temperature, and used this trait to find that the Kurly protein may be involved in initiating movement rather than keeping the cilia moving once they’ve started.
The team also explored proteins that interact with Kurly. The Northwestern team showed that when the kur gene was inactivated using a gene-editing technique called CRISPR-Cas9, the lack of a functioning Kurly protein led to the mis-positioning of a second protein on the cell surface called Prickle2, which helps cells know which direction they face. Without proper Prickle2 positioning, the cilia pushed fluid in the wrong direction.
The study of the Kurly protein involved Grimes as well as two additional co-authors, Kimberly Jaffe and Jodi Schottenfeld-Roames, a former postdoctoral researcher and graduate student respectively, in the Burdine lab. The initial studies on the Kurly protein were conducted as part of an undergraduate research project by Tse-shuen (Jade) Ku, Class of 2007. Additional work was contributed by Nicholas Morante and José Pelliccia, graduate students in the Burdine lab.
The work at Northwestern University was performed in the laboratory of Brian Mitchell with the assistance of Michael Werner and Sun Kim.
The research was supported by a National Institutes of Health (NIH) Ruth L. Kirschstein Institutional National Research Service Award grant to K. Jaffe (#1F32HD060396-01A1), an NIH National Institute of General Medical Sciences grant to B. Mitchell (#2R01GM089970), and an NIH Eunice Kennedy Shriver National Institute of Child Health and Human Development grant to R. Burdine (#2R01HD048584).
Kimberly M. Jaffe, Daniel T. Grimes, Jodi Schottenfeld-Roames, Michael E. Werner, Tse-Shuen J. Ku, Sun K. Kim, Jose L. Pelliccia, Nicholas F.C. Morante, Brian J. Mitchell, Rebecca D. Burdine.c21orf59/kurly controls both cilia motility and polarization. Cell Reports (2016), http://dx.doi.org/10.1016/j.celrep.2016.01.069. In Press Corrected Proof.


You must be logged in to post a comment.