By Tien Nguyen, Department of Chemistry
A fully extended strand of human DNA measures about five feet in length. Yet it occupies a space just one-tenth of a cell by wrapping itself around histones—spool-like proteins—to form a dense hub of information called chromatin.
Access to these meticulously packed genes is regulated by post-translational modifications, chemical changes to the structure of histones that act as on-off signals for gene transcription. Mistakes or mutations in histones can cause diseases such as glioblastoma, a devastating pediatric brain cancer.
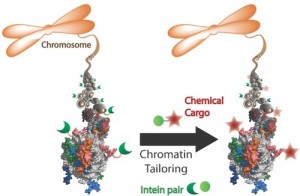
Researchers at Princeton University have developed a facile method to introduce non-native chromatin into cells to interrogate these signaling pathways. Published on April 6 in the journal Nature Chemistry, this work is the latest chemical contribution from the Muir lab towards understanding nature’s remarkable information indexing system.
Tom Muir, the Van Zandt Williams, Jr. Class of ’65 Professor of Chemistry, began investigating transcriptional pathways in the so-called field of epigenetics almost a decade earlier. Deciphering such a complex and dynamic system posed a formidable challenge, but his research lab was undeterred. “It’s better to fail at something important than to succeed at something trivial,” he said.
Muir recognized the value of introducing chemical approaches to epigenetics to complement early contributions that came mainly from molecular biologists and geneticists. If epigenetics was like a play, he said, molecular biology and genetics could identify the characters but chemistry was needed to understand the subplots.
These subplots, or post-translational modifications of histones, of which there are more than 100, can occur cooperatively and simultaneously. Traditional methods to probe post-translational modifications involved synthesizing modified histones one at a time, which was a very slow process that required large amounts of biological material.
Last year, the Muir group introduced a method that would massively accelerate this process. The researchers generated a library of 54 nucleosomes—single units of chromatin, like pearls on a necklace—encoded with DNA-barcodes, unique genetic tags that can be easily identified. Published in the journal Nature Methods, the high throughput method required only microgram amounts of each nucleosome to run approximately 4,500 biochemical assays.
“The speed and sensitivity of the assay was shocking,” Muir said. Each biochemical assay involved treatment of the DNA-barcoded nucleosome with a writer, reader or nuclear extract, to reveal a particular binding preference of the histone. The products were then isolated using a technique called chromatin immunoprecipitation and characterized by DNA sequencing, essentially an ordered readout of the nucleotides.
“There have been incredible advances in genetic sequencing over the last 10 years that have made this work possible,” said Manuel Müller, a postdoctoral researcher in the Muir lab and co-author on the Nature Methods article.
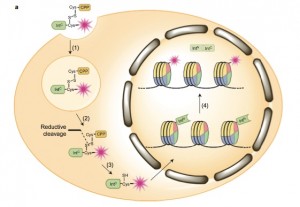
With this method, researchers could systematically interrogate the signaling system to propose mechanistic pathways. But these mechanistic insights would remain hypotheses unless they could be validated in vivo, meaning inside the cellular environment.
The only method for modifying histones in vivo was extremely complicated and specific, said Yael David, a postdoctoral researcher in the Muir lab and lead author on the recent Nature Chemistry study that demonstrated a new and easily customizable approach.
The method relied on using ultra-fast split inteins, protein fragments that have a great affinity for one another. First, one intein fragment was attached to a modified histone, by encoding it into a cell. Then, the other intein fragment was synthetically fused to a label, which could be a small protein tag, fluorophore or even an entire protein like ubiquitin.
Within minutes of being introduced into the cell, the labeled intein fragment bound to the histone intein fragment. Then like efficient and courteous matchmakers, the inteins excised themselves and created a new bond between the label and modified histone. “It’s really a beautiful way to engineer proteins in a cell,” David said.
Regions of the histone may be loosely or tightly packed, depending on signals from the cell indicating whether or not to transcribe a gene. By gradually lowering the amount of labeled intein introduced, the researchers could learn about the structure of chromatin and tease out which areas were more accessible than others.
Future plans in the Muir lab will employ these methods to ask specific biological questions, such as whether disease outcomes can be altered by manipulating signaling pathway. “Ultimately, we’re developing methods at the service of biological questions,” Muir said.
This research was supported by the US National Institutes of Health (grants R37-GM086868 and R01 GM107047).
Read the articles:
Nguyen, U.T.T.; Bittova, L.; Müller, M.; Fierz, B.; David, Y.; Houck-Loomis, B.; Feng, V.; Dann, G.P.; Muir, T.W. “Accelerated chromatin biochemistry using DNA-barcoded nucleosome libraries.” Nature Methods, 2014, 11, 834.
David, Y.; Vila-Perelló, M; Verma, S.; Muir, T.W. “Chemical tagging and customizing of cellular chromatin states using ultrafast trans-splicing inteins.” Nature Chemistry, Advance online publication, April 6, 2015.

You must be logged in to post a comment.