By Catherine Zandonella, Office of the Dean for Research
A new study has identified genes involved in long-term memory in the worm as part of research aimed at finding ways to retain cognitive abilities during aging.
The study, which was published in the journal Neuron, identified more than 750 genes involved in long-term memory, including many that had not been found previously and that could serve as targets for future research, said senior author Coleen Murphy, an associate professor of molecular biology and the Lewis-Sigler Institute for Integrative Genomics at Princeton University.
“We want to know, are there ways to extend memory?” Murphy said. “And eventually, we would like to ask, are there compounds that could maintain memory with age?”
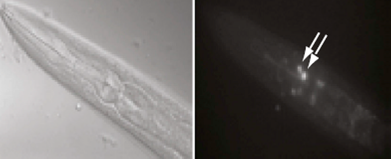
The newly pinpointed genes are “turned on” by a molecule known as CREB (cAMP-response element-binding protein), a factor known to be required for long-term memory in many organisms, including worms and mice.
“There is a pretty direct relationship between CREB and long-term memory,” Murphy said, “and many organisms lose CREB as they age.” By studying the CREB-activated genes involved in long-term memory, the researchers hope to better understand why some organisms lose their long-term memories as they age.
To identify the genes, the researchers first instilled long-term memories in the worms by training them to associate meal-time with a butterscotch smell. Trained worms were able to remember that the butterscotch smell means dinner for about 16 hours, a significant amount of time for the worm.
The researchers then scanned the genomes of both trained worms and non-trained worms, looking for genes turned on by CREB.
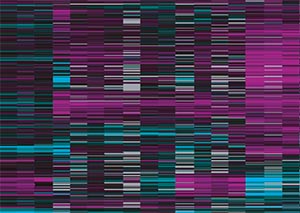
The researchers detected 757 CREB-activated genes in the long-term memory-trained worms, and showed that these genes were turned on primarily in worm cells called the AIM interneurons.
They also found CREB-activated genes in non-trained worms, but the genes were not turned on in AIM interneurons and were not involved in long-term memory. CREB turns on genes involved in other biological functions such as growth, immune response, and metabolism. Throughout the worm, the researchers noted distinct non-memory (or “basal”) genes in addition to the memory-related genes.
The next step, said Murphy, is to find out what these newly recognized long-term memory genes do when they are activated by CREB. For example, the activated genes may strengthen connections between neurons.
Worms are a perfect system in which to explore that question, Murphy said. The worm Caenorhabditis elegans has only 302 neurons, whereas a typical mammalian brain contains billions of the cells.
“Worms use the same molecular machinery that higher organisms, including mammals, use to carry out long-term memory,” said Murphy. “We hope that other researchers will take our list and look at the genes to see whether they are important in more complex organisms.”
Murphy said that future work will involve exploring CREB’s role in long-term memory as well as reproduction in worms as they age.
The team included co-first-authors Postdoctoral Research Associate Vanisha Lakhina, Postdoctoral Research Associate Rachel Arey, and Associate Research Scholar Rachel Kaletsky of the Lewis-Sigler Institute for Integrative Genomics. Additional research was performed by Amanda Kauffman, who earned her Ph.D. in Molecular Biology in 2010; Geneva Stein, who earned her Ph.D. in Molecular Biology in 2014; William Keyes, a laboratory assistant in the Department of Molecular Biology; and Daniel Xu, who earned his B.A. in Molecular Biology in 2014.
Funding for the research was provided by the National Institutes of Health and the Paul F. Glenn Laboratory for Aging Research at Princeton University.
Citation: Vanisha Lakhina, Rachel N. Arey, Rachel Kaletsky, Amanda Kauffman, Geneva Stein, William Keyes, Daniel Xu, and Coleen T. Murphy. Genome-wide Functional Analysis of CREB/Long-Term Memory-Dependent Transcription Reveals Distinct Basal and Memory Gene Expression Programs, Neuron (2015), http://dx.doi.org/10.1016/j.neuron.2014.12.029.

You must be logged in to post a comment.