By: Tien Nguyen
Bacteria speak to one another using peptide signals in a soundless language known as quorum sensing. In a step towards translating bacterial communications, researchers at Princeton University have revealed the structure and biosynthesis of streptide, a peptide involved in the quorum sensing system common to many streptococci.

“It’s extremely rare for one research group to do both natural products discovery and mechanistic enzymology,” said Leah Bushin, a member of the Seyedsayamdost lab and co-first author on the article published on April 20 in Nature Chemistry. Bushin worked on elucidating the structure of streptide as part of her undergraduate senior thesis project and will enter Princeton Chemistry’s graduate program in the fall.
To explore how bacteria communicate, first she had to grow them, a challenging process in which oxygen had to be rigorously excluded. Next she isolated the streptide and analyzed it using two-dimensional (2D) nuclear magnetic resonance (NMR) spectroscopy, a technique that allows scientists to deduce the connections between atoms in a molecule by pulsing their nuclei with powerful magnets to pulse atomic nuclei.
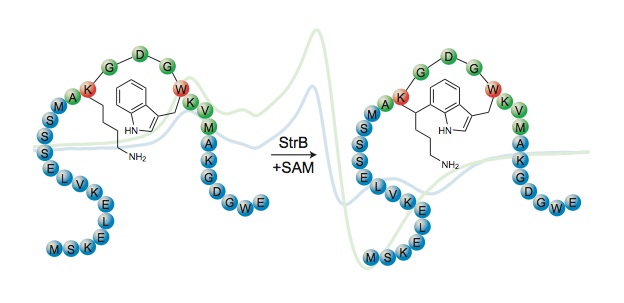
The experiments revealed that streptide contained an unprecedented crosslink between two unactivated carbons on lysine and tryptophan, constituting a new class of macrocyclic peptides. “We didn’t think it would be as cool as a carbon-carbon bond between two amino acid side chains, so it was definitely a surprise.” said Bushin.
To figure out how this novel bond was being formed, the researchers took a closer look at the gene cluster that produced streptide. Within the gene cluster, they suspected a radical S-adenosyl methionine (SAM) enzyme, which they dubbed StrB, could be responsible for this unusual modification.

“Radical SAM enzymes catalyze absolutely amazing chemistries,” said Kelsey Schramma, a graduate student in the Seyedsayamdost lab and co-first author on the article. “There are over 48,000 radical SAM enzymes, but only about 50 have been characterized and just a dozen or so studied in detail,” she said.
To probe the enzyme’s role in making streptide, the researchers created a mutated version of the bacteria lacking the strB gene. The mutant failed to produce streptide, confirming that the StrB enzyme was significant and warranted further study.
Schramma determined that in order to function properly, the StrB enzyme required some key components: the pre-crosslinked substrate, which she prepared synthetically, cofactor SAM, reductant, and two iron-sulfur (Fe-S) clusters carefully assembled in the protein interior. The team then showed that one of the FeS clusters reductively activated one molecule of SAM, kicking off a chain of one-electron (radical) reactions that gave rise to the novel carbon-carbon bond.
“The synergy between Leah and Kelsey was great,” said Mohammad Seyedsayamdost, an assistant professor of chemistry at Princeton who led the research team. “They expressed interest in complementary aspects of the project and the whole ended up being greater than the sum of its parts,” he said.
Their efforts included not only chemical and biological approaches, but also theoretical computational studies. While the 2D NMR experiments revealed the flat structure of streptide, its three-dimensional conformation was still unknown.
“Since the crosslink had never been reported, we had to code the modification into the program, which took a bit of creativity,” Bushin said. After corresponding with the software creator, they were able to confidently assign a key residue in the macrocycle with the S-configuration.
Future work will target streptide’s biological function—its meaning in the bacterial language—as well as confirming its production by other streptococcal bacteria strains.
“What we have revealed is a new and unusual mechanism that nature uses to synthesize macrocyclic peptides. There is a lot of novel chemistry to be discovered by interrogating bacterial secondary metabolite biosynthetic pathways,” Seyedsayamdost said.
Read the article here:
Schramma, K. R.; Bushin, L. B.; Seyedsayamdost, M. R. “Structure and biosynthesis of a macrocyclic peptide containing an unprecedented lysine-to-tryptophan crosslink.” Nature Chemistry, 2015, 7, 431.
This work was supported by the National Institutes of Health (grant no. GM098299), and by Princeton University start-up funds.

You must be logged in to post a comment.