By Kristin Qian for the Office of the Dean for Research
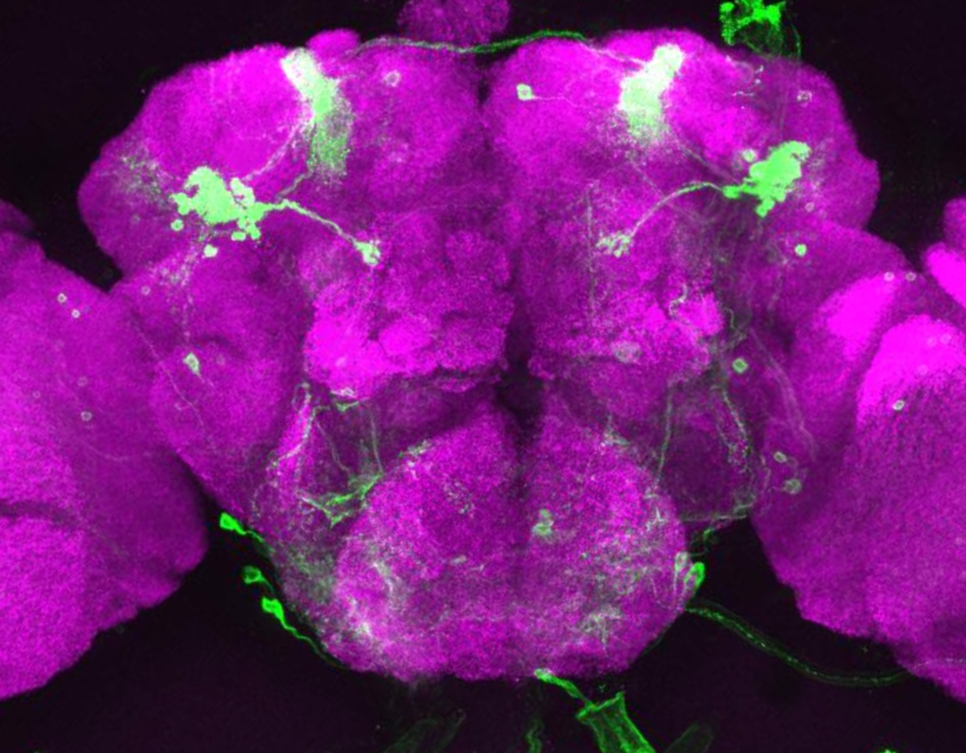
Researchers at Princeton University have developed a highly sensitive and precise method to explore genes important for memory formation within single neurons of the Drosophila fly brain. With this method, the researchers found an unexpected result: certain genes involved in creating long-term memories in the brain are the same ones that the eye uses for sensing light.
The study, published in the May 17 issue of the journal Cell Reports, demonstrated the utility of the new method and also identified new patterns of gene expression that drive long-term memory formation.
“Ultimately, to understand the brain, we want to know what individual neurons are doing,” said Mala Murthy, assistant professor in the Princeton Neuroscience Institute and the Department of Molecular Biology. “We found that single neurons can be defined by their pattern of their gene expression, even if they’re all in the same brain network.”
To their surprise, the researchers found that many of the active genes in these neurons produce proteins that are best known for their roles in detecting light in the fly’s eye or sensing odor in the fly’s nose. “It is possible that these sensory proteins have been repurposed by the brain for a different function,” Murthy said.
“Even though the paper is focused on the methodology, which I think will be impactful for the field, there is this new science here—a whole new class of molecules we found that is in the central brain and seems to be involved in memory formation,” Murthy said.
Researchers have known that genes “turn on,” or start making proteins, during the formation of long-term memories in Drosophila, a widely used organism in studies of neurobiology, but they didn’t know exactly which genes in which neurons were involved.
To investigate this question, the researchers first trained flies to form long-term memories. Then they extracted single neurons from the fly brains and evaluated all of the gene readouts, or transcripts, which encode proteins. By comparing the transcripts of the memory-trained flies to those of non-trained flies, researchers were able to identify genes involved in long-term memory formation.
The task was complicated by the tiny size of the fly’s head, which is just one millimeter across, and contains fewer than 100,000 neurons. Murthy’s team focused on neuron types in one part of the brain, the mushroom body, named for its distinctive shape.
First author Amanda Crocker, a former postdoctoral fellow in Murthy’s lab and now an assistant professor of neuroscience at Middlebury College, conducted the experiments in collaboration with co-authors Xiao-Juan Guan, a senior research specialist in the Princeton Neuroscience Institute; Coleen Murphy, professor of molecular biology and the Lewis-Sigler Institute for Integrative Genomics; and Murthy.
“Our work opens up the ability to use Drosophila as a way to study how gene expression in single neurons relates to brain function,” Crocker said. “This has been a challenge because the fly brain is very small and contains fewer neurons than other organisms that neuroscientists study. The advantage of using flies is that they have significantly less redundancy in the neurons that they do have. We can look at specific neurons and gene expression, and ask what the genes are doing in that cell to cause the behavior.”
The researchers trained the flies to form long-term memories by exposing them to an odor – either an earthy, mushroom-like smell (3-octanol) or a menthol-like smell (4-methylcyclohexanol) – while simultaneously delivering a negative stimulus in the form of an electric shock.
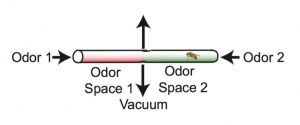
The training took place in a tube containing the two odors, one at each end of the tube. Researchers paired one of the odors with the electric shock, and as a result the fly avoided that end of the tube. The assay was conducted in the dark, so that the flies could use only their sense of smell, not their vision, to navigate the tube.
A second group of flies received the electric shock and the odor, but not at the same time, so they did not form the memory that linked odor to shock.
The researchers then isolated single neurons from the fly brains using tiny glass tubes to suction out the cells. Harvesting neurons using this technique is not common, Murthy said, and it had not been combined with a complete analysis of gene activity in fly neurons before. With this novel method, they were able to use only 10 to 90 femtograms – a quintillionth of a kilogram – of genetic material.
They evaluated gene activity by looking at the production of messenger ribonucleic acid (mRNA), an intermediary between DNA and proteins. The result is a “transcriptome,” or readout of all of the genetic messages that the cell uses to produce proteins. The researchers then read the transcriptome to see which genes produced proteins in the memory-trained flies versus the non-trained flies, and found that some of the active genes in memory-trained flies were the same as ones used in the sensory organs to detect light, odors and taste.
To follow-up, the researchers bred mutant flies that lacked genes for some of the light-sensing proteins and thus could not see. The same memory experiments as before were carried out, and the researchers confirmed that the flies lacking light-sensing proteins were both unable to see and unable to form long-term memories.
The discovery of the expression of genes for classical ‘light-sensing’ proteins, such as rhodopsin, as well as other sensory-related proteins for odor and taste detection, was unexpected because these proteins were not known to be utilized in mushroom bodies, Murthy said. Although studies in other organisms, including humans, have detected sensory genes in areas of the brain unrelated to the sensory organ itself, this may be the first study to link these genes to memory formation.
The study was funded by a National Institutes of Health Ruth L. Kirschstein Institutional National Research Service Award, the Alfred P. Sloan Foundation, the Human Frontier Science Program, a National Science Foundation (NSF) CAREER award, the McKnight Endowment Fund for Neuroscience, the Klingenstein Foundation, a National Institutes of Health New Innovator award, and an NSF BRAIN Initiative EAGER award. The study was also funded in part through Princeton’s Glenn Center for Quantitative Aging Research, directed by Coleen Murphy.
The paper, “Cell-Type-Specific Transcriptome Analysis in the Drosophila Mushroom Body Reveals Memory-Related Changes in Gene Expression,” was published in the May 17 issue of Cell Reports.

You must be logged in to post a comment.