By Morgan Kelly, Office of Communications

Stopping the outbreak of a disease hinges on a wealth of data such as what makes a suitable host and how a pathogen spreads. But gathering these data can be difficult for diseases in remote areas of the world, or for epidemics involving wild animals.
A new study led by Princeton University researchers and published in the Journal of the Royal Society Interface explores an approach to studying epidemics for which details are difficult to obtain. The researchers analyzed the 2013 outbreak of dolphin morbillivirus — a potentially fatal pathogen from the same family as the human measles virus — that resulted in more than 1,600 bottlenose dolphins becoming stranded along the Atlantic coast of the United States by 2015. Because scientists were able to observe dolphins only after they washed up on shore, little is known about how the disease transmits and persists in the wild.
The researchers used a Poisson process — a statistical tool used to model the random nature of disease transmission — to determine from sparse data how dolphin morbillivirus can spread. They found that individual bottlenose dolphins may be infectious for up to a month and can spread the disease over hundreds of miles, particularly during seasonal migrations. In 2013, the height of disease transmission occurred toward the end of summer around an area offshore of Virginia Beach, Virginia, where multiple migratory dolphin groups are thought to cross paths.
In the interview below, first author Sinead Morris, a graduate student in ecology and evolutionary biology, explains what the researchers learned about the dolphin morbillivirus outbreak, and how the Poisson process can help scientists understand human epidemics. Morris is in the research group of co-author Bryan Grenfell, Princeton’s Kathryn Briger and Sarah Fenton Professor of Ecology and Evolutionary Biology and Public Affairs.
Q: How does the Poisson process track indirectly observed epidemics and what specific challenges does it overcome?
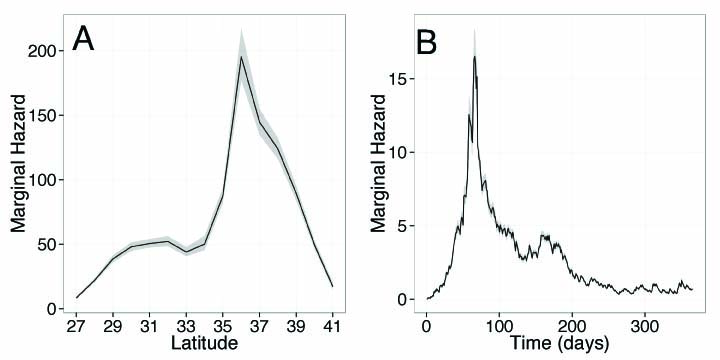
A: One of the main challenges in modeling indirectly observed epidemics is a lack of data. In our case, we had information on all infected dolphins that had been found stranded on shore, but had no data on the number of individuals that became infected but did not strand. The strength of the Poisson process is that its simple framework means it can be used to extract important information about the how the disease is spreading across space and time, despite having such incomplete data. Essentially the way the process works is that it keeps track of where and when individual dolphins stranded, and then at each new point in the epidemic it uses the history of what has happened before to project what will happen in the future. For example, an infected individual is more likely to transmit the disease onwards to other individuals in close spatial proximity than to those far away. So, by keeping track of all these infections the model can identify where and when the largest risk of new infections will be.
Q: Why was this 2013-15 outbreak of dolphin morbillivirus selected for study, and what key insights does this work provide?
A: The recent outbreak of dolphin morbillivirus spread rapidly along the northwestern Atlantic coast from New York to Florida, causing substantial mortality among coastal bottlenose dolphin populations. Despite the clear detrimental impact that this disease can have, however, it is still poorly understood. Therefore, our aim in modeling the epidemic was to gain much needed information about how the virus spreads. We found that a dolphin may be infectious for up to 24 days and can travel substantial distances (up to 220 kilometers, or 137 miles) within this time. This is important because such long-range movements — for example, during periods of seasonal migration — are likely to create many transmission opportunities from infected to uninfected individuals, and may have thus facilitated the rapid spread of the virus down the Atlantic coast.
Q: Can this model be used for human epidemics?
A: The Poisson process framework was originally developed to model the occurrence of earthquakes, and has since been used in a variety of other contexts that also tend to suffer from noisy, indirectly observed data, such as urban crime distribution. To model dolphin morbillivirus, we adapted the framework to incorporate more biological information, and similar techniques have also been applied to model meningococcal disease in humans, which can cause meningitis and sepsis. Generally, the data characterizing human epidemics are more detailed than the data we had for this project and, as such, models that can incorporate greater complexity are more widely used. However, we hope that our methods will stimulate the greater use of Poisson process models in epidemiological systems that also suffer from indirectly observed data.
This research was supported by the RAPIDD program of the Science and Technology Directorate of the Department of Homeland Security; the National Institutes of Health Fogarty International Center; the Bill and Melinda Gates Foundation; and the Marine Mammal Unusual Mortality Event Contingency Fund and John H. Prescott Marine Mammal Rescue Assistance Grant Program operated by the National Oceanic and Atmospheric Administration.
Sinead E. Morris, Jonathan L. Zelner, Deborah A. Fauquier, Teresa K. Rowles, Patricia E. Rosel, Frances Gulland and Bryan T. Grenfell. “Partially observed epidemics in wildlife hosts: modeling an outbreak of dolphin morbillivirus in the northwestern Atlantic, June 2013–2014.” Journal of the Royal Society Interface, published Nov. 18 2015. DOI: 10.1098/rsif.2015.0676

You must be logged in to post a comment.