By Catherine Zandonella, Office of the Dean for Research
Serendipity – the act of finding something good or useful while not specifically searching for it – can sometimes pay off. Now Princeton University chemistry researchers report that this non-specific type of searching has yielded a new method of building molecules for use in new drugs, new agricultural chemicals and even new perfumes.
In a paper published today in the journal Science, Princeton’s David MacMillan and his team describe the discovery of a new chemical reaction – not noted before in nature or in any lab – that could assist pharmaceutical chemists and others who routinely create new chemicals for a variety of industries.
Until now, no one realized this chemical reaction – which involves adding atoms to a specific carbon atom on a molecule – could occur, according to MacMillan, the James S. McDonnell Distinguished University Professor of Chemistry at Princeton. “If you show this chemical reaction to most chemists, they immediately say ‘that’s impossible,'” MacMillan said.
In this case, the team discovered this “impossible” reaction using an approach MacMillan pioneered that he calls “accelerated serendipity.” The researchers use robotic arms to conduct thousands of reactions per day by combining in test tubes different combinations of chemicals along with catalysts that spur the reactions. When the investigators find a reaction that makes an interesting product, they study it to understand how the reaction occurs.
“We didn’t invent this new reaction – nature did that,” MacMillan said, “but we figured out how to get the reaction to happen in the lab.” said MacMillan. His team, which included graduate student Michael Pirnot, postdoctoral researcher David Martin and former postdoctoral researcher Danica Rankic, uses ordinary light bulbs as catalysts, a technique developed in MacMillan’s lab and published in Science in 2008, to spur the reactions.
Going forward, chemists can add this new reaction to their tool box of methods for building up molecules, which they do in a way analogous to joining together pieces of Kinex or Tinker Toys, by swapping in new parts to increase the function of the molecule. In the new reaction published today, the team discovered a way to join so-called “functional groups” to a specific carbon atom (see diagram) in larger structures known as ketones and aldehydes. The ability to add functional groups to that carbon atom was thought impossible until now.
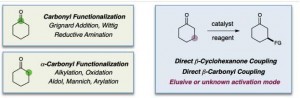
This new chemical reaction has wide applications, MacMillan said. “This is a fundamental reaction which any chemist can start using.”
For example, a chemist who is building a drug to treat Alzheimer’s disease might desire to add a chemical group to the reluctant carbon atom. Normally that would require the chemist to conduct several different chemical reactions over several weeks, but with the new reaction the chemist could build the drug in two days and be testing drug candidates much more quickly.
Similarly a chemist at a fragrance company could use the new reaction to experiment with the creation of new perfume formulations.
MacMillan’s original paper on accelerated serendipity, published in 2011 in Science, successfully discovered a reaction now used in the drug industry. Yet it was controversial because other scientists interpreted the robotic searches as random searches, when in fact they were not random. “We chose chemicals that had never been shown to react with each other – those are the ones we believe might lead to as-yet undiscovered reactions.” MacMillan said that these reactions may have been created in the past by chemists who didn’t recognize what they were.
Michael T. Pirnot, Danica A. Rankic, David B. C. Martin, David W. C. MacMillan. Photoredox Activation for the Direct β-Arylation of Ketones and Aldehydes. Science 29 March 2013. Vol. 339 no. 6127 pp. 1593-1596.
This research was supported by the National Institute of General Medical Sciences grant R01 GM103558-01 and gifts from Merck, Amgen, Abbott, and Bristol-Myers Squibb.


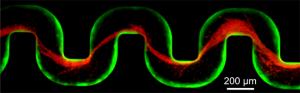 A new study has examined how bacteria clog medical devices, and the result isn’t pretty. The microbes join to create slimy ribbons that tangle and trap other passing bacteria, creating a full blockage in a startlingly short period of time.
A new study has examined how bacteria clog medical devices, and the result isn’t pretty. The microbes join to create slimy ribbons that tangle and trap other passing bacteria, creating a full blockage in a startlingly short period of time.
You must be logged in to post a comment.