Our blog has moved to become a part of Discovery: Research at Princeton. Visit us in our new home.
Experimental treatment shows promise against triple-negative breast cancer in mouse study
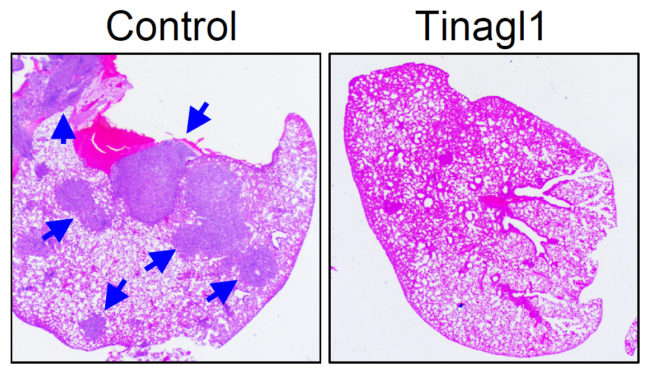
By simultaneously tackling two mechanisms for cancer’s growth, an experimental therapy reduced the spread of triple-negative breast cancer in a study conducted in mice.
The therapeutic agent, called Tinagl1, is based on a naturally occurring protein and blocks two of the main pathways by which breast cancer cells can grow and migrate out of the primary tumor to spread to other organs in the body. The study was published in the journal Cancer Cell on Jan. 3.
“People have tried to block the spread of this form of cancer but attempts so far have failed because if you try one approach, the cancer cells compensate by finding a way to escape,” said Yibin Kang, the Warner-Lambert/Parke-Davis Professor of Molecular Biology at Princeton University, associate director for consortium research at the Rutgers Cancer Institute of New Jersey, and senior author on the study. “With this new approach, the treatment blocks both pathways at the same time. It is like having one stone that kills two birds.”
Triple-negative breast cancer is a highly aggressive form of cancer that occurs in 12 to 17 percent of all breast cancer cases. It gets its name from the lack of three prominent biological targets – the estrogen receptor (ER), the progesterone receptor (PR), and the human epidermal growth factor receptor 2 (HER2) – that are used to find and kill cancer cells.
Patients with triple-negative breast cancer have high rates of recurrence and fewer treatment options along with the likelihood of the cancer developing resistance to treatments. This leads to an overall worse prognosis for patients.
The new findings suggests that recombinant Tinagl1, is a good candidate for further exploration and development toward a treatment for this form of cancer.
The researchers found that Tinagl1 inhibited two main pathways that contribute to triple-negative breast cancer’s aggressiveness and ability to resist treatments. Tinagl1 inhibited both pathways in distinct ways, overcoming the compensation mechanisms that the cancer uses to evade treatments.
Tinagl1 tackles cancer by stopping the action of a tumor-promoting protein known as epidermal growth factor receptor gene (EGFR). Mutations in this gene cause EGFR signaling activity to spike, sending pro-growth signals to cells and fueling tumor growth and metastasis to new regions of the body. Treatments that target EGFR have had limited clinical success, perhaps because the cancer cells find new pathways for growth.
The protein also has effects on molecules called integrins that are involved in regulating cellular migration to new locations, adhesion to other cells, and transformation into tumors. Tinagl1 appears able to target integrin signaling by interfering with a related protein called focal adhesion kinase (FAK), which promotes cell migration, growth and survival.
The two pathways are molecularly interconnected, causing redundancies and compensations that contribute to the aggressiveness and treatment-resistance of triple-negative breast cancer, according to the researchers.
The researchers looked at over 800 breast tumor samples taken from human patients. They found that tumor samples that had lower expression of the Tinagl1 gene were more likely to have come from patients with advanced tumor stages and shorter survival times. Tumors with higher levels of the gene were more strongly linked to good patient outcomes. The difference was most prominent among the triple negative subgroup of breast cancers.
To test whether having the Tinagl1 gene could protect against tumor growth and spread, the researchers engineered human and mouse tumor cells to express high levels of the Tinagl1 gene. The researchers found that high expression of Tinagl1 in mouse cancer cells produced slower-growing tumors that were less likely to metastasize to the lung.
The researchers also administered Tinagl1 protein to mice with mammary tumors and found that treatment for seven weeks significantly inhibited primary tumor growth and spontaneous lung metastasis, while having no significant side effects. In a study to look at the timing of Tinagl1 treatment, the compound was effective even after tumors had begun to metastasize. The team also investigated the mechanism of Tinagl1 suppression and found that the protein inhibits both EGFR and integrin/FAK signaling pathways, thus achieving a better outcome than using single inhibitors against each of these two pathways.
The study was led by the first author Minhong Shen, a postdoctoral research associate; with Princeton co-authors including associate professional specialist Yong Wei; former graduate student Brian Ell; graduate student Xinlei Sheng; postdoctoral research associate Mark Esposito; former undergraduate researcher Jooeun Kang, Class of 2014; research specialist Xiang Hang; postdoctoral fellow Hanqiu Zheng; undergraduate Michelle Rowicki, Class of 2020; postdoctoral fellow Toni Celia-Terrassa; Professor of Molecular Biology Ileana Cristea; and Yibin Kang. Additional contributors were Yi-Zhou Jiang, Yirong Liu and Zhi-Ming Shao at Fudan University Shanghai Cancer Center; Weichung Shih at the Rutgers Cancer Institute of New Jersey; and Lanjing Zhang of the University Medical Center of Princeton.
Funding for the study was provided by the Brewster Foundation, a grant from the U.S. Department of Defense (BC123187) and postdoctoral fellowships from Susan G. Komen (PDF17332118) and the New Jersey Commission on Cancer Research (DFHS15PPCO21).
Vaccine signatures in humanized mice point to better understanding of infectious diseases
Infectious diseases kill millions of people each year, but the search for treatments is hampered by the fact that laboratory mice are not susceptible to some human viruses, including killers like human immunodeficiency virus (HIV). For decades, researchers have turned to mice whose immune systems have been “humanized” to respond in a manner similar to humans.
Now a team at Princeton University has developed a comprehensive way to evaluate how immune responses of humanized mice measure up to actual humans. The research team looked at the mouse and human immune responses to one of the strongest vaccines known, a yellow fever vaccine called YFV-17D. The comparison of these “vaccine signatures” showed that a newly developed humanized mouse developed at Princeton shares significant immune-system responses with humans. The study was published in the journal Nature Communications.
“Understanding immune responses to human pathogens and potential vaccines remains challenging due to differences in the way our human immune system responds to stimuli, as compared to for example that of conventional mice, rats or other animals,” said Alexander Ploss, associate professor of molecular biology at Princeton. “Until now a rigorous method for testing the functionality of the human immune system in such a model has been missing. Our study highlights an experimental paradigm to address this gap.”
Humanized mice have been used in infectious disease research since the late 1980s. Yet without rigorous comparisons, researchers know little about how well the mice predict human responses such as the production of infection-fighting cells and antibodies.
To address this issue, researchers exposed the mice to the YFV-17D vaccine, which is made from a weakened, or attenuated, living yellow fever virus. Vaccines protect against future infection by provoking the production of antibodies and immune-system cells.
In previous work, the researchers explored the effect of YFV-17D on conventional humanized mice. But the researchers found that the mice responded only weakly. This led them to develop a mouse with responses that are more similar to those of humans.
To do so, the researchers introduced additional human genes for immune system components — such as molecules that detect foreign invaders and chemical messengers called cytokines — so that the complexity of the engrafted human immune system reflected that of humans. They found that the new mice have responses to YFV-17D that are very similar to the responses seen in humans. For example, the pattern of gene expression that occurs in response to YFV-17D in the mice shared significant similarities to that of humans. This signature gene expression pattern, reflected in the “transcriptome,” or total readout of all of the genes of the organism, translated into better control of the yellow fever virus infection and to immune responses that were more specific to yellow fever.
The researchers also looked at two other types of immune responses: the cellular responses, involving production of cytotoxic T cells and natural killer cells that attack and kill infected cells, and the production of antibodies specific to the virus. By evaluating these three types of responses – transcriptomic, cellular, and antibody – in both mice and humans, the researchers produced a reliable platform for evaluating how well the mice can serve as proxies for humans.
Florian Douam, a postdoctoral research associate and the first author on the study, hopes that the new testing platform will help researchers explore exactly how vaccines induce immunity against pathogens, which in many cases is not well understood.
“Many vaccines have been generated empirically without profound knowledge of how they induce immunity,” Douam said. “The next generation of mouse models, such as the one we introduced in our study, offer unprecedented opportunities for investigating the fundamental mechanisms that define the protective immunity induced by live-attenuated vaccines.”
Mice bearing human cells or human tissues have the potential to aid research on treatments for many diseases that infect humans but not other animals, such as – in addition to HIV – Epstein Barr Virus, human T-cell leukemia virus, and Karposi sarcoma-associated herpes virus.
“Our study highlights the importance of human biological signatures for guiding the development of mouse models of disease,” said Ploss. “It also highlights a path toward developing better models for human immune responses.”
The study involved contributions from Florian Douam, Gabriela Hrebikova, Jenna Gaska, Benjamin Winer and Brigitte Heller in Princeton University’s Department of Molecular Biology; Robert Leach, Lance Parsons and Wei Wang in Princeton University’s Lewis Sigler Institute for Integrative Genomics; Bruno Fant at the University of Pennsylvania; Carly G. K. Ziegler and Alex K. Shalek of Massachusetts Institute of Technology and Harvard Medical School; and Alexander Ploss in Princeton University’s Department of Molecular Biology.
The research was supported the National Institutes of Health (NIH, R01AI079031 and R01AI107301, to A.P) and an Investigator in Pathogenesis Award by the Burroughs Wellcome Fund (to A.P.). Additionally, A.K.S. was supported by the Searle Scholars Program, the Beckman Young Investigator Program, the NIH (1DP2OD020839, 5U24AI118672, 1U54CA217377, 1R33CA202820, 2U19AI089992, 1R01HL134539, 2RM1HG006193, 2R01HL095791, P01AI039671), and the Bill & Melinda Gates Foundation (OPP1139972). C.G.K.Z. was supported by a grant from the National Institute of General Medical Sciences (NIGMS, T32GM007753). J.M.G. and B.Y.W. were supported by a pre-doctoral training grant from the NIGMS (T32GM007388). B.Y.W. was also a recipient of a pre-doctoral fellowship from the New Jersey Commission on Cancer Research.
By Catherine Zandonella, Office of the Dean for Research
Climate warming to boost major hurricanes in active Atlantic seasons

New NOAA research that looks at the devastating 2017 Atlantic hurricane season projects that if similar weather conditions occur in the future, it’s likely that the number of major hurricanes (category 3 and higher) would increase by two in a similar active year at the end of century.
This increase would be driven by predicted climate warming, according to the research appearing today in Science.
The finding was reported by a team at NOAA’s Geophysical Fluid Dynamics Laboratory (GFDL) that included lead author Hiroyuki Murakami, who conducted the research while an associate research scholar with the Program in Atmospheric and Oceanic Sciences at Princeton University
Last year’s six major Atlantic hurricanes included landfalling hurricanes Harvey, Irma and Maria that unleashed destruction on and caused loss of life in communities across Texas, Florida and Puerto Rico. The three hurricanes caused an estimated $265 billion in damages during a year that shattered all records for U.S. economic losses due to severe weather.
Global model identifies climate influence

Using a high-resolution global climate model called HiFLOR, developed at GFDL, scientists were able to accurately predict the active hurricane season in June 2017. Scientists then conducted additional experiments with HiFLOR that found it was a remarkably warm tropical Atlantic Ocean, relative to the global tropical ocean, which was the main driver of 2017 hurricane activity.
“This new method allows us to predict hurricane activity as the season is happening as well as take into consideration the likely contribution of climate warming,” said Murakami, a climate researcher at GFDL. NOAA scientists use ocean temperature data showing the relative warmth of the tropical Atlantic to help create vital hurricane season outlooks.
In addition to Murakami, the research team included Emma Levin of Paul D. Schreiber High School in Port Washington, NY, Thomas Delworth and Richard Gudgel of GFDL, and Pang-Chi Hsu of Nanjing University of Information Science and Technology, China.
The study, “Dominant Effect of Relative Tropical Atlantic Warming on Major Hurricane Occurrence,” by H. Murakami, E. Levin, T. L. Delworth, R. Gudgel and P-C Hsu, was published in the journal Science on Sept. 27, 2018.
Funding for this study was provided by the National Oceanic and Atmospheric Administration (NOAA).
Article courtesy of NOAA.
The notorious Luminous Blue Variable star

New, three-dimensional simulations reveal the inner workings of one of the universe’s most mysterious stars.
Sparkling with an exceptional blue-toned brilliance and exhibiting wild variations in both brightness and spectrum, the luminous blue variable (LBV) is a relatively rare and still somewhat mysterious type of star.
Video courtesy of the University of California-Santa BarbaraIts appearance tends to fluctuate radically over time, and that has piqued the curiosity of astrophysicists who wonder what processes may be at play.
“The luminous blue variable is a supermassive, unstable star,” said Yan-Fei Jiang, a researcher at the University of California-Santa Barbara’s Kavli Institute for Theoretical Physics (KITP) who earned his doctorate at Princeton University in 2013.
Jiang is the first author on a new study published in the journal Nature this week. The paper’s senior author is Princeton’s James Stone, the Lyman Spitzer Jr., Professor of Theoretical Astrophysics, Stone, a professor of astrophysical sciences and applied and computational mathematics, and the chair of Princeton’s Department of Astrophysical Sciences. Jiang and Stone developed the numerical code used in the study.
Unlike our own comparatively smaller and steady-burning Sun, LBVs have been shown to burn bright and hot, then cool and fade so as to be almost indistinguishable from other stars, only to flare up again. Because of these changes, conventional one-dimensional models have been less than adequate at explaining the special physics of these stars.
However, thanks to special, data-intensive supercomputer modeling conducted at Argonne National Laboratory’s Argonne Leadership Computing Facility (ALCF) for its INCITE program, Jiang and colleagues — Matteo Cantiello, a visiting associate research scholar in Princeton’s Department of Astrophysical Sciences, and an associate research scientist at the Simons Foundation’s Flatiron Institute; Lars Bildsten of KITP; Eliot Quataert at UC Berkeley; Omer Blaes of UCSB; and Stone of Princeton — have now developed a three-dimensional simulation. It not only shows the stages of an LBV as it becomes progressively more luminous, then erupts, but also depicts the physical forces that contribute to that behavior. The simulation was developed also with computational resources from NASA and the National Energy Research Scientific Computing Center.
Of particular interest to the researchers are the stars’ mass loss rates, which are significant compared to those of less massive stars. Understanding how these stellar bodies lose mass, Jiang said, could lead to greater insights into just how they end their lives as bright supernova.
Among the physical processes never before seen with one-dimensional models are the supersonic turbulent motions — the ripples and wrinkles radiating from the star’s deep envelope as it prepares for a series of outbursts.
“These stars can have a surface temperature of about 9,000 degrees Kelvin during these outbursts,” Jiang said. That translates to 15,740 degrees Fahrenheit or 8,726 degrees Celsius.
Also seen for the first time in three dimensions is the tremendous expansion of the star immediately before and during the outbursts — phenomena not captured with previous one-dimensional models. The three dimensional simulations show that it is the opacity of the helium that sets the observed temperature during the outburst.
According to Jiang, in a one-dimensional stellar evolution code, helium opacity — the extent to which helium atoms prevent photons (light) from escaping — is not very important in the outer envelope because the gas density at the cooler outer envelope is far too low.
The paper’s co-author and KITP Director Lars Bildsten explained that the three-dimensional model demonstrates that “the region deep within the star has such vigorous convection that the layers above that location get pushed out to much larger radii, allowing the material at the location where helium recombines to be much denser.” The radiation escaping from the star’s hot core pushes on the cooler, opaque outer region to trigger dramatic outbursts during which the star loses large amounts mass. Hence, convection — the same phenomena responsible for thundercloud formation — causes not only variations in the star’s radius but also in the amount of mass leaving in the form of a stellar wind.
Additional work is underway on more simulations, according to Jiang, including models of the same stars but with different parameters such as metallicity, rotation and magnetic fields.
“We are trying to understand how these parameters will affect the properties of the stars,” Jiang said. “We are also working on different types of massive stars — the so-called Wolf-Rayet stars — which also show strong mass loss.”
The journal article is “Luminous Blue Variable Outbursts from the Variations of Helium Opacity,” by Yan-Fei Jiang, Matteo Cantiello, Lars Bildsten, Eliot Quataert, Omer Blaes and James Stone. Nature, September 27, 2018.
This story was provided by the University of California-Santa Barbara.
This research was supported in part by the NASA ATP grant ATP-80NSSC18K0560, the National Science Foundation under grant number NSF PHY 11-25915, 17-48958, and in part by a Simons Investigator award from the Simons Foundation (to E.Q.) and the Gordon and Betty Moore Foundation through grant GBMF5076. An award of computer time was provided by the Innovative and Novel Computational Impact on Theory and Experiment (INCITE) program. This research used resources of the Argonne Leadership Computing Facility and National Energy Research Scientific Computing Center, which are DOE Offices of Science User Facility supported under contract DE-AC02-06CH11357 and DE-AC02-05CH11231. Resources supporting this work were also provided by the NASA High-End Computing (HEC) program through the NASA Advanced Supercomputing (NAS) Division at Ames Research Center. The Flatiron Institute is supported by the Simons Foundation
How rabies virus moves through nerve cells, and how it might be stopped
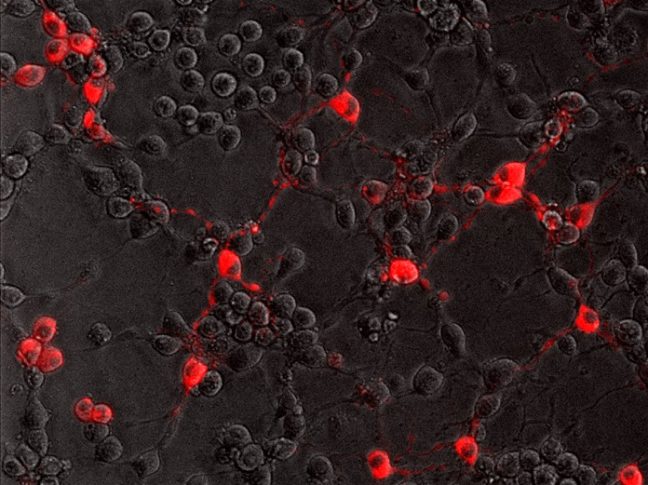
To successfully infect its host, the rabies virus must move from the nerve ending to the nerve cell body where it can replicate. In a study published July 20 in the journal PLoS Pathogens, researchers from Princeton University reveal that the rabies virus moves differently compared to other neuron-invading viruses and that its journey can be blocked by a drug commonly used to treat amoebic dysentery.
Most viruses only infect the nervous system accidentally when the immune system is compromised. But some “neurotropic” viruses have evolved to target neurons as part of their normal infectious cycle. The rabies virus, for example, is transmitted when an infected animal bites into a host’s muscle. It then spreads into the end terminals of motor neurons innervating the muscle and travels along the neurons’ long axon fibers to the neuronal cell bodies. From there, the virus can spread throughout the central nervous system and into the salivary glands, where it can be readily transmitted to other hosts. Though rabies infections in humans are rare in the United States, the virus kills nearly 60,000 people annually.
Alpha herpesviruses, such as herpes simplex viruses, also enter peripheral nerve terminals and move along axons to the neuronal cell body, where they can lie dormant for the life of the host.
“Transport to the neuronal cell body is not a passive process, but an active one relying on the neuron’s own motor proteins and microtubule tracks,” said Lynn Enquist, Princeton’s Henry L. Hillman Professor in Molecular Biology, a professor of molecular biology and the Princeton Neuroscience Institute, and the study’s senior author. “Virus particles must engage this machinery for efficient transport in axons, otherwise infection cannot start.”
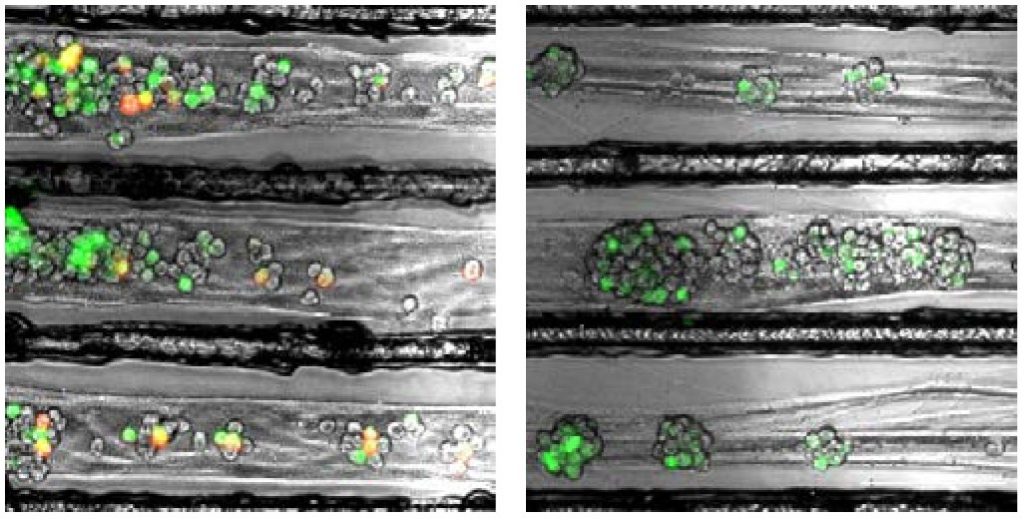
Enquist and colleagues previously found that alpha herpesviruses engage the neuronal transport machinery by stimulating protein synthesis at infected nerve terminals. Viral transport to the cell body can therefore be blocked by drugs that inhibit protein synthesis, as well as by cellular antiviral proteins called interferons.
In the current study, Enquist and colleagues investigated how the rabies virus engages the neuronal transport machinery. The researchers infected neurons with a virulent strain of the virus tagged with a red fluorescent protein, allowing the researchers to observe viral transport in real time by live-cell fluorescence microscopy.
The study was led by Margaret MacGibeny, who earned her Ph.D. in 2018, and associate research scholar Orkide Koyuncu, at Princeton, with contributions from research associate Christoph Wirblich and Matthias Schnell, professor and chair of microbiology and immunology at Thomas Jefferson University.
In contrast to alpha herpesvirus infections, the team found that interferons had no effect on rabies virus transport, perhaps because, until it reaches the neuronal cell body, the rabies virus hides out inside cellular structures called endosomes.
“We also couldn’t detect increased protein synthesis in axons upon rabies virus infection,” MacGibeny said. “But, to our surprise, we saw that a protein synthesis inhibitor called emetine efficiently blocked rabies virus transport to the cell body.”
Emetine had no effect on the transport of endosomes devoid of the rabies virus. But endosomes carrying the virus were either completely immobilized, or were only able to move short distances at slower-than-normal speeds.
Other protein synthesis inhibitors did not block rabies virus transport, however, suggesting that emetine works by inhibiting a different process in infected neurons.
“Emetine has been used to treat amoebic dysentery,” Koyuncu said. “In the laboratory it is widely used to inhibit protein synthesis but there are recent reports indicating that emetine has anti-viral effects that are independent of protein synthesis inhibition. Our study shows that this drug can inhibit rabies virus invasion of the nervous system through a novel mechanism that hasn’t been reported before.”
“The manuscript by MacGibeny et al. both advances and complicates our understanding of how neurotropic viruses make their way from the axon terminus to the cell body,” said Professor Glenn Rall, an expert in neurotropic virus infections at Fox Chase Cancer Center, who was not involved in the study. “Revealing variations in the axonal transport of neurotropic viruses, coupled with intriguing insights into new roles for well-known drugs, has both mechanistic and clinical implications for these life-threatening infections.
“Our next step is to figure out how emetine disrupts rabies virus transport in axons,” Enquist says. “Does it inhibit cell signaling pathways after rabies virus entry, or does it directly block the recruitment of motor proteins to virus-carrying endosomes?”
This study was funded by the US National Institutes of Health (grants P40 OD010996, RO1 NS33506, and F30 NS090640).
The study, Retrograde axonal transport of rabies virus is unaffected by interferon treatment but blocked by emetine locally in axons, by Margaret A. MacGibeny, Orkide O. Koyuncu, Christoph Wirblich, Matthias J. Schnell, and Lynn W. Enquist was published in PLoS Pathogens. 2018. DOI: 10.1371/journal.ppat.1007188
Text courtesy of the Department of Molecular Biology
A valley so low: Electrons congregate in ways that may be useful to “valleytronics”
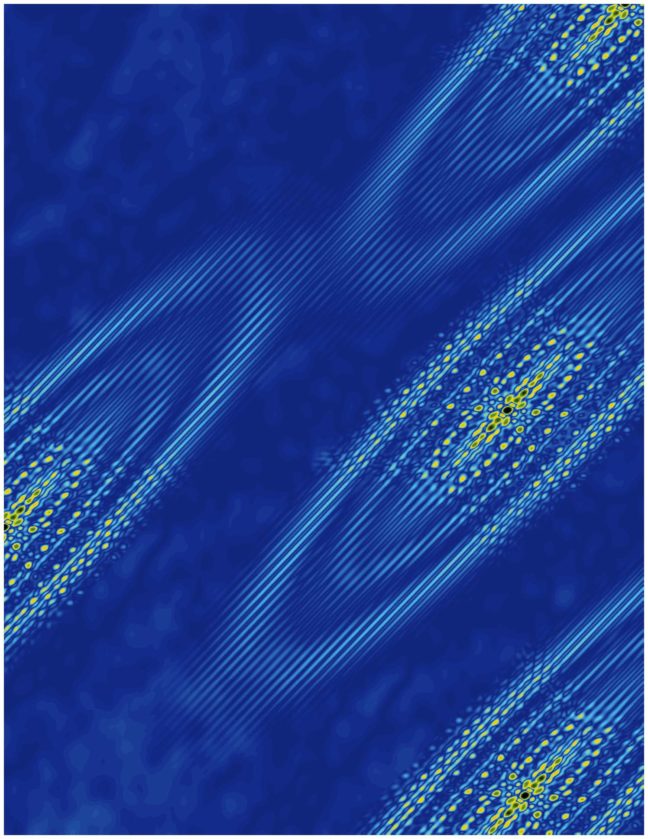
A Princeton University-led study has revealed an emergent electronic behavior on the surface of bismuth crystals that could lead to insights on the growing area of technology known as “valleytronics.”
The term refers to energy valleys that form in crystals and that can trap single electrons. These valleys potentially could be used to store information, greatly enhancing what is capable with modern electronic devices.
In the new study, researchers observed that electrons in bismuth prefer to crowd into one valley rather than distributing equally into the six available valleys. This behavior creates a type of electricity called ferroelectricity, which involves the separation of positive and negative charges onto opposite sides of a material. The study was published in the journal Nature Physics.
The finding confirms a recent prediction that ferroelectricity arises naturally on the surface of bismuth when electrons collect in a single valley. These valleys are not literal pits in the crystal but rather are like pockets of low energy where electrons prefer to rest.
The researchers detected the electrons congregating in the valley using a technique called scanning tunneling microscopy, which involves moving an extremely fine needle back and forth across the surface of the crystal. They did this at temperatures hovering close to absolute zero and under a very strong magnetic field, up to 300,000 times greater than Earth’s magnetic field.
The behavior of these electrons is one that could be exploited in future technologies. Crystals consist of highly ordered, repeating units of atoms, and with this order comes precise electronic behaviors. Silicon’s electronic behaviors have driven modern advances in technology, but to extend our capabilities, researchers are exploring new materials. Valleytronics attempts to manipulate electrons to occupy certain energy pockets over others.
The existence of six valleys in bismuth raises the possibility of distributing information in six different states, where the presence or absence of an electron can be used to represent information. The finding that electrons prefer to cluster in a single valley is an example of “emergent behavior” in that the electrons act together to allow new behaviors to emerge that wouldn’t otherwise occur, according to Mallika Randeria, the first author on the study and a graduate student at Princeton working in the laboratory of Ali Yazdani, the Class of 1909 Professor of Physics.
“The idea that you can have behavior that emerges because of interactions between electrons is something that is very fundamental in physics,” Randeria said. Other examples of interaction-driven emergent behavior include superconductivity and magnetism.
In addition to Randeria, the study included equal contributions from Benjamin Feldman, a former postdoctoral fellow at Princeton who is now an assistant professor of physics at Stanford University, and Fengcheng Wu, a postdoctoral researcher at Argonne National Laboratory. Additional contributors at Princeton were Hao Ding, a postdoctoral research associate in physics, and András Gyenis, a postdoctoral research associate in electrical engineering; Ji Huiwen, who earned a doctoral degree at Princeton and is now a postdoctoral researcher at the University of California-Berkeley; Robert Cava, Princeton’s Russell Wellman Moore Professor of Chemistry; and Yazdani. Additional contributions came from Allan MacDonald, professor of physics at the University of Texas-Austin.
The study was funded by the Gordon and Betty Moore Foundation as part of the EPiQS initiative (GBMF4530), the U.S. Department of Energy (DOE-BES grant DE-FG02-07ER46419), the U.S. Army Research Office MURI program (W911NF-12-1-046), the National Science Foundation’s MRSEC program through the Princeton Center for Complex Materials (NSF-DMR-142054 and NSF-DMR-1608848), and the Eric and Wendy Schmidt Transformative Technology Fund at Princeton. Work at University of Texas-Austin was supported by DOE grant (DE-FG03-02ER45958) and by the Welch Foundation (TBF1473).
The study “Ferroelectric quantum Hall phase revealed by visualizing Landau level wave function interference,” by Mallika T. Randeria, Benjamin E. Feldman, Fengcheng Wu, Hao Ding, András Gyenis, Huiwen Ji, R. J. Cava, Allan H. MacDonald, and Ali Yazdani, was published online May 14, 2018, and in print in August, 2018, in the journal Nature Physics.
By Catherine Zandonella
Genetically engineered mice could boost fight against aggressive hepatitis
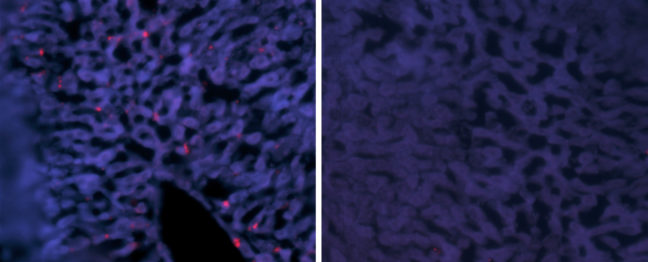
Article provided by the Department of Molecular Biology
Hepatitis delta virus (HDV) causes the most aggressive form of viral hepatitis in humans, putting at least 20 million people worldwide at risk of developing liver fibrosis, cirrhosis, and liver cancer. Efforts to develop effective treatments against HDV have been hampered by the fact that laboratory mice are not susceptible to the virus. But, in a study published June 27, 2018, in the journal Science Translational Medicine, Alexander Ploss, assistant professor of molecular biology at Princeton University and colleagues describe a genetically engineered mouse that can be persistently infected with HDV.
HDV is a small, RNA-based “satellite” virus that produces just a single protein of its own and therefore requires additional proteins provided by another liver virus, hepatitis B virus (HBV). HDV can infect patients already carrying HBV, or both viruses can infect patients simultaneously. Though infections can be prevented with an anti-HBV vaccine, there are no antiviral therapies available to cure existing HDV infections.
HDV and HBV infect the liver by binding to a protein called NTCP that is present on the surface of liver cells. But the viruses only recognize the version of NTCP present in humans and a few other primates, and therefore can’t infect mice or other small mammals that produce their own versions of NTCP. This has made it difficult to study HBV and HDV infections in the laboratory. Researchers have tried transplanting human liver cells into immunocompromised mice before infecting them with virus, but this approach has produced inconsistent results and is both expensive and time-consuming.
Ploss and colleagues, led by graduate student Benjamin Winer, took a different approach. They generated mice that express the human NTCP protein in their liver cells, allowing these cells to be infected by HBV and HDV.
In these mice, HBV failed to replicate after entering mouse liver cells but HDV was able to establish persistent infection when provided with the HBV proteins it needs to propagate. For example, mice genetically engineered to produce both human NTCP and the entire HBV genome could be infected with HDV for up to 14 days. “To our knowledge, this is the first time the entire HDV life cycle has been recapitulated in a mouse model with inheritable susceptibility to HDV,” Ploss said.
The mice were able to rid themselves of HDV before they developed any liver damage, apparently by mounting an immune response involving antiviral interferon proteins and various white blood cell types, including Natural Killer (NK) cells and T cells. Accordingly, mice expressing human NTCP and the HBV genome, but lacking functional B, T, and NK cells could be infected with HDV for two months or more.
These immunocompromised animals allowed Ploss and colleagues to test the effectiveness of two drugs that are currently being developed as treatments for HDV infection. Both drugs—either alone or in combination—suppressed the levels of HDV in immunocompromised mice after viral infection. But the drugs were not able to completely clear the mice of HDV; viral levels rose again within weeks of stopping treatment.
“This is largely in line with recently reported data from clinical trials, showing the utility of our model for preclinical antiviral drug testing,” Winer said.
“Our model is amenable to genetic manipulations, robust, and can be adopted as a method to rapidly screen for potential treatments,” Ploss added.
Timothy M. Block, president of the Hepatitis B Foundation and its Baruch S. Blumberg Institute who was not involved in the study, said “These systems should be able to provide practical, and presumably economical tools. Their work is urgently needed, and a desperate community welcomes it. I emphasize that it is often the new methods in science that revolutionize a field such as drug discovery, almost as much as the new drugs themselves.”
The research team included collaborators from Princeton University; Weill Medical College of Cornell University; The Jackson Laboratory; University Medical Center Hamburg-Eppendorf, Hamburg; New York University Medical Center; and North Carolina State University College of Veterinary Medicine.
This study was supported by grants from the National Institutes of Health (R01 AI079031, R01 AI107301, R21AI117213 to Alexander Ploss), a Research Scholar Award from the American Cancer Society (RSG-15-048-01-MPC to Alexander Ploss), a Burroughs Wellcome Fund Award for Investigators in Pathogenesis (to Alexander Ploss) and a Graduate fellowship from the Health Grand Challenge from the Global Health Fund of Princeton University (to Benjamin Y. Winer). The NYU Experimental Pathology Immunohistochemistry Core Laboratory is supported in part by the Laura and Isaac Perlmutter Cancer Center Support Grant; NIH/NCI P30CA016087 and the National Institutes of Health S10 Grants NIH/ORIP S10OD01058 and S10OD018338. Benjamin Y. Winer is a recipient of F31 NIH/NRSA Ruth L. Kirschstein Predoctoral awarded from the NIAID. Julie Sellau and Elham Shirvani-Dastgerdi are both recipients of postdoctoral fellowships from the German Research Foundation. Michael V. Wiles was funded by The Jackson Laboratory.
Benjamin Y. Winer, Elham Shirvani-Dastgerdi, Yaron Bram, Julie Sellau, Benjamin E. Low, Heath Johnson, Tiffany Huang, Gabriela Hrebikova, Brigitte Heller, Yael Sharon, Katja Giersch, Sherif Gerges, Kathleen Seneca, Mihai-Alexandru Pais, Angela S. Frankel, Luis Chiriboga, John Cullen, Ronald G. Nahass, Marc Lutgehetmann, Jared Toettcher, Michael V. Wiles, Robert E. Schwartz, and Alexander Ploss. Preclinical assessment of antiviral combination therapy in a genetically humanized mouse model for persistent hepatitis delta virus infection. Science Translational Medicine. 2018. DOI: 10.1126/scitranslmed.aap9328
Novel insulators with conducting edges
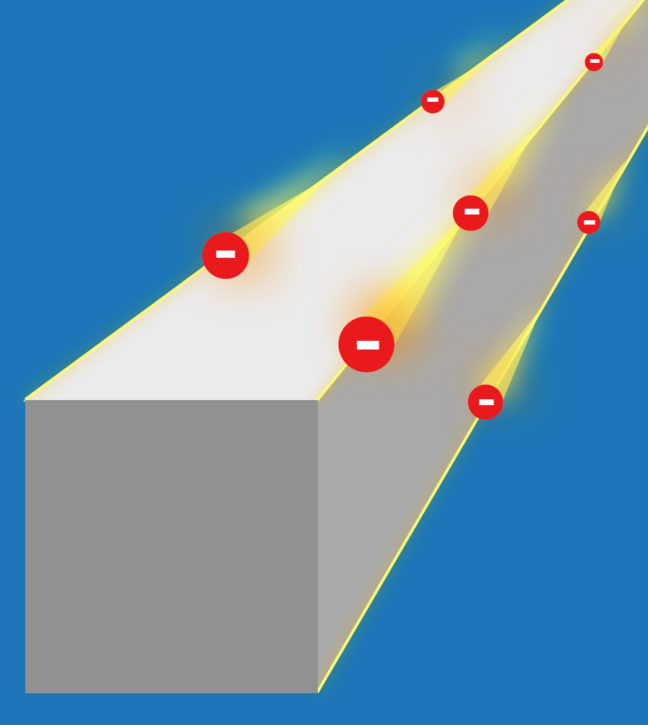
Article courtesy of the University of Zurich
An international team including scientists at Princeton University is researching a new class of materials: higher-order topological insulators. The edges of these crystalline solids conduct electric current without dissipation, while the rest of the crystal remains insulating. These materials could be useful for applications in semiconductor technology and for building quantum computers. The study was published in the journal Sciences Advances.
Topology examines the properties of objects and solids that are protected against perturbations and deformations. Topological materials known so far include topological insulators, which are crystals that insulate on the inside but conduct electrical current on their surface. The conducting surfaces are topologically protected, which means that they cannot easily be brought into an insulating state.
A new class of materials: Higher-order topological insulators
Theoretical physicists have now predicted a new class of topological insulators that have conducting properties on the edges of crystals rather than on the surface. The research team, made up of scientists from University of Zurich (UZH), Princeton University, the Donostia International Physics Center, and the Max Planck Institute of Microstructure Physics in Halle, dubbed the new material class “higher-order topological insulators.” The extraordinary robustness of the conducting edges makes them particularly interesting: The current of topological electrons cannot be stopped by disorder or impurities. If an imperfection gets in the way of the current, it simply flows around the impurity.
Like a highway for electrons
In addition, the crystal edges do not have to be specially prepared to conduct electrical current. If the crystal breaks, the new edges also conduct current. “The most exciting aspect is that electricity can at least in theory be conducted without any dissipation,” said Titus Neupert, professor in the Department of Physics at UZH. “You could think of the crystal edges as a kind of highway for electrons. They can’t simply make a U-turn.” Neupert and his team collaborated with B. Andrei Bernevig, professor of physics, and his team at Princeton University.
This property of dissipationless conductance, more commonly associated with superconductors at low temperatures, is not shared with the previously known topological insulator crystals that have conducting surfaces, but is specific to the higher-order topological crystals.
Further theoretical and experimental research needed
The physicists’ study still mostly relies on theoretical aspects. They have proposed tin telluride as the first compound to show these novel properties. “More material candidates have to be identified and probed in experiments,” says Neupert. The researchers hope that in the future, nanowires made of higher-order topological insulators may be used as conducting paths in electric circuits. These nanowires could be combined with magnetic and superconducting materials for use in building quantum computers.
Funding was provided by the U.S. Department of Energy (DE-SC0016239), a Simons Investigator Award, the David and Lucile Packard Foundation, and the Eric and Wendy Schmidt Transformative Technology Fund at Princeton University. The computational part of the Princeton work was performed with funding from the National Science Foundation’s Early-Concept Grants for Exploratory Research (DMR-1643312) and Materials Research Science and Engineering Centers (DMR-1420541), the Office of Naval Research (ONR-N00014-14-1-0330), and the Army Research Office (MURI W911NF-12-1-0461). Support was also provided by the Swiss National Science Foundation (grant number: 200021_169061), the European Union’s Horizon 2020 research and innovation program (ERC-StG-Neupert-757867-PARATOP), and the Spanish Ministry of Economy and Competitiveness (FIS2016-75862-P).
The study, “Higher-order topological insulators,” by Frank Schindler, Ashley M. Cook, Maia G. Vergniory, Zhijun Wang, Stuart S. P. Parkin, B. Andrei Bernevig, Titus Neupert was published online in Science Advances, on June 1st, 2018. DOI: 10.1126/sciadv.aat0346.
Half a degree more global warming could flood out 5 million more people

By Liz Fuller-Wright, Office of Communications
The 2015 Paris climate agreement sought to stabilize global temperatures by limiting warming to well below 2.0 degrees Celsius above pre-industrial levels and to pursue limiting warming even further, to 1.5 C.
To quantify what that would mean for people living in coastal areas, a group of researchers employed a global network of tide gauges and a local sea level projection framework to explore differences in the frequency of storm surges and other extreme sea-level events across three scenarios: global temperature increases of 1.5, 2.0 and 2.5 C.

They concluded that by 2150, the seemingly small difference between an increase of 1.5 and 2.0 C would mean the permanent inundation of lands currently home to about 5 million people, including 60,000 who live on small island nations.
The study, conducted by researchers at Princeton University and colleagues at Rutgers and Tufts Universities, the independent scientific organization Climate Central, and ICF International, was published in the journal Environmental Research Letters on March 15, 2018.
“People think the Paris Agreement is going to save us from harm from climate change, but we show that even under the best-case climate policy being considered today, many places will still have to deal with rising seas and more frequent coastal floods,” said D.J Rasmussen, a graduate student in Princeton’s Program in Science, Technology and Environmental Policy in the Woodrow Wilson School of Public and International Affairs, and first author of the study.
The researchers found that higher temperatures will make extreme sea level events much more common. They used long-term hourly tide gauge records and extreme value theory to estimate present and future return periods of extreme sea-level events through the 22nd century. Under the 1.5 C scenario, the frequency of extreme sea level events is still expected to increase. For example, by the end of the 21st century, New York City is expected to experience one Hurricane Sandy-like flood event every five years.
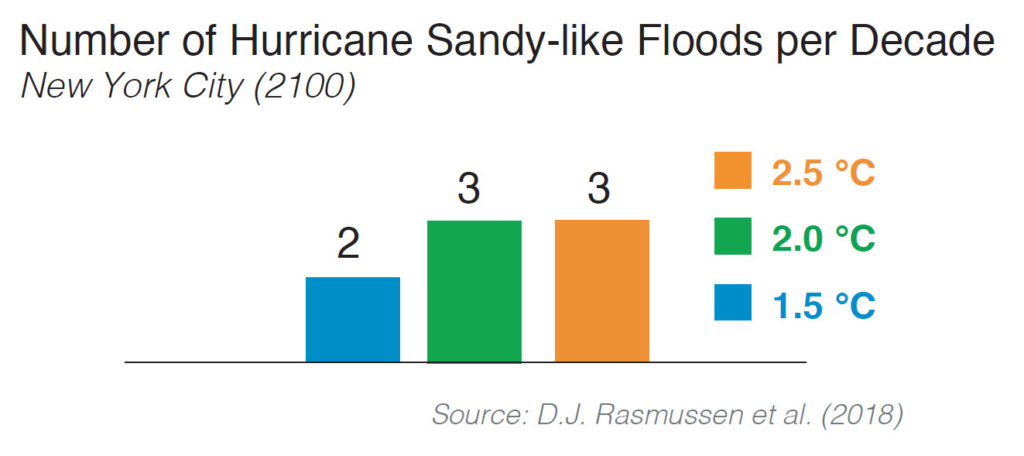
Extreme sea levels can arise from high tides or storm surge or a combination of surge and tide (sometimes called the storm tide). When driven by hurricanes or other large storms, extreme sea levels flood coastal areas, threatening life and property. Rising mean sea levels are already magnifying the frequency and severity of extreme sea levels, and experts predict that by the end of the century, coastal flooding may be among the costliest impacts of climate change in some regions.
Future extreme events will be exacerbated by the rising global sea level, which in turn depends on the trajectory of global mean surface temperature. Even if global temperatures are stabilized, sea levels are expected to continue to rise for centuries, due to the fact that carbon dioxide stays in the atmosphere for a long time and the ice sheets are slow to respond to warming.
Overall, the researchers predicted that by the end of the century, a 1.5 C temperature increase could drive the global mean sea level up by roughly 1.6 feet (48 cm) while a 2.0 C increase will raise oceans by about 1.8 feet (56 cm) and a 2.5 C increase will raise sea level by an estimated 1.9 feet (58 cm).
The research team included Klaus Bittermann, a postdoctoral researcher at Tufts University who is associated with the Potsdam Institute for Climate Impact Research in Germany; Maya Buchanan, who earned a doctorate in 2017 from the Woodrow Wilson School of Public and International Affairs and is now at ICF International; Scott Kulp, a senior computational scientist and senior developer for the Program on Sea Level Rise at Climate Central, an independent organization of scientists and journalists located in Princeton, NJ; Benjamin Strauss, who received a Ph.D. in 2007 from Princeton’s Department of Ecology and Evolutionary Biology and is chief scientist at Climate Central; and Robert Kopp, a professor of Earth and Planetary Sciences at Rutgers University.
The senior author on the study was Michael Oppenheimer, the Albert G. Milbank Professor of Geosciences and International Affairs and the Princeton Environmental Institute. Kopp developed the sea level projection framework, and Buchanan, Kopp, and Oppenheimer developed the flood projection framework. Researchers at Climate Central conducted the population inundation analysis.
The research was supported by the National Science Foundation (grants EAR-1520683 and ICER-1663807), the Rhodium Group as part of the Climate Impact Lab consortium, and NASA (grant 80NSSC17K0698).
“Extreme sea level implications of 1.5 °C, 2.0 °C, and 2.5 °C temperature stabilization targets in the 21st and 22nd centuries” by D.J Rasmussen, Klaus Bittermann, Maya Buchanan, Scott Kulp, Benjamin Strauss, Robert Kopp, and Michael Oppenheimer was published March 15, 2018 in Environmental Research Letters. (doi: 10.1088/1748-9326/aaac87).

You must be logged in to post a comment.